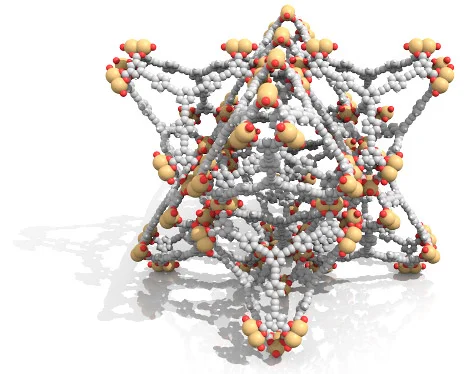
The above screenshots are taken from the opening part of an animation named High Density Energy Storage Using Self-Assembled Materials created by Christopher Wilmer and his colleagues. The animation was one of the winners of 2011 NSF International Science & Engineering Visualization Challenge. Starting from a single metal ion (blue), we see organic molecules (white/grey) attached to metal ions, forming sophisticated 3D structures. The camera zooms out, revealing macroscopic blue crystals made of these organic molecules and metal ions. And finally we see a futuristic car which is powered by natural gas stored inside these blue crystals. You can watch the full animation below:
Chris is currently an assistant professor in the Department of Chemical & Petroleum Engineering at the University of Pittsburgh, having earned his Ph.D. in Chemical and Biological Engineering from Northwestern University. He is also the co-founder and advisor of NuMat Technologies, a cleantech company that computationally designs and synthesizes high performing nanomaterials for gas storage and separation applications. An expert in advanced computational modeling, Chris was named to the “30 under 30 in Energy” list at Forbes Magazine. He was additionally honored as a “top solver” by InnoCentive, being amongst the top 16 individuals out of over 200,000 globally in the submission of innovative solutions to real-world challenges.
I am thrilled to show his work on L2Molecule. Also I am grateful he took the time to answer a few questions about scientific visualization.
Q & A with Christopher Wilme
1. What are your favorite tools for creating scientific visualizations?
My favorite tool is Blender, and I try to use it for everything. The movie "High Density Energy Storage Using Self-assembled Materials" was done entirely using Blender. Sometimes I am pressed for time and so I use more specialized tools, like Visual Molecular Dynamics (VMD). When I need to make a compelling diagram in 5 minutes or less, I use Microsoft Powerpoint, which unbeknownst to many actually lets you build surprisingly sophisticated 3D structures. Other programs I use are Mathematica and 3D Coat. Of course I also use Gimp and Inkscape for simple image manipulation.
2. What were your roles in making the above animation and how long did it take to create it?
The three "directors" were Dr. Omar Farha, Patrick Fuller, and myself. At the time, I was the only one with any experience in animation, but Pat took this opportunity to learn and made several contributions to the animation (including the scene corresponding to the attached image). We came up with the idea for the animation shortly before the deadline for the NSF Visualization Challenge, and so the whole thing (animation, music, narration, editing) was done in three weeks. The research depicted in the movie was carried out primarily by Omar and myself, and so we crafted the narrative together from which I made a typical "story board" before animating anything. I wore all of the hats one could think of in the process: doing the underlying scientific research, conceiving the idea and narrative, script writing, modeling, animating, sequencing, organizing the contributions of others, etc. The only thing I played no part in was composing the music for the third scene, which was done by my brother, Alex Wilmer.
Nature Chemistry cover created by Christopher Wilmer.
Nu-100, a new metal-organic framework material. Image created by Christopher Wilmer.
3. What is the most challenging part in making this animation?
The tools for making high quality animations are not very user friendly or intuitive (at least not the ones I use). If you use them frequently, perhaps because animation is your day job, it's not as much of a problem. For someone like myself, who makes animations like this more sporadically, it's frustrating to keep relearning some of techniques. To combat this, starting next year I will be teaching a course called "Scientific Visual Communication" which will codify a lot of the techniques I have used, which will hopefully help my students (and me!) produce animations like this without so much head scratching.
4. There is a strong connection between the first part of your animation and a scene in Stanley Kubrick’s 2001: A Space Odyssey. Can you tell us a little bit about this?
For many years I have dreamed of making an animation depicting molecular self-assembly as an elegant nanoscale dance, much like the satellites and docking space vessels in Kubrick's movie. The catalyst for this animation, the NSF Visualization Challenge, provided the perfect opportunity to make that dream a reality. In many ways, the analogy between docking space ships and self-assembling molecules is a good one. Both processes are awe-inspiring, and as I argue in the animation, they each leverage extreme technological precision. The engineering and physics utilized to launch satellites into precise orbits is mirrored by the chemistry that is needed to grow macroscopic crystals that are (nearly) perfect in their atomic detail. Someday, when science has advanced further and we are able to routinely make synthetic molecular machines, I might make another Kubrick inspired movie.
5. In your opinion, how important is creating engaging visuals for scientific research?
It's more important than people think, even when taking this sentence into account. Very few people will admit to not being interested in- or not paying attention to- a scientific presentation simply because it isn't visually engaging. However, many people (students and professors alike) have come up to me after my presentations to remark that they were especially engaged because of the animations. It falls on the shoulders of scientists to best communicate their results to the outside world; we shouldn't expect others to decipher our cryptic research if it hasn't been communicated well.
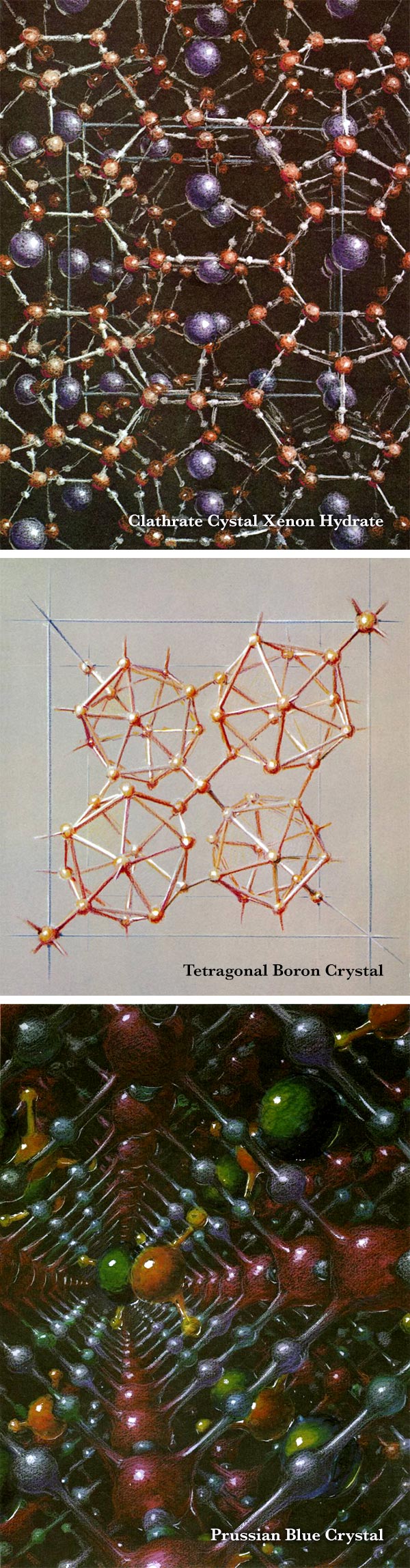
This visualization shows the absorption of methane molecules (orange spheres) inside different MOF (metal-organic framework) materials. Image created by Christopher Wilmer and Patrick Fuller.